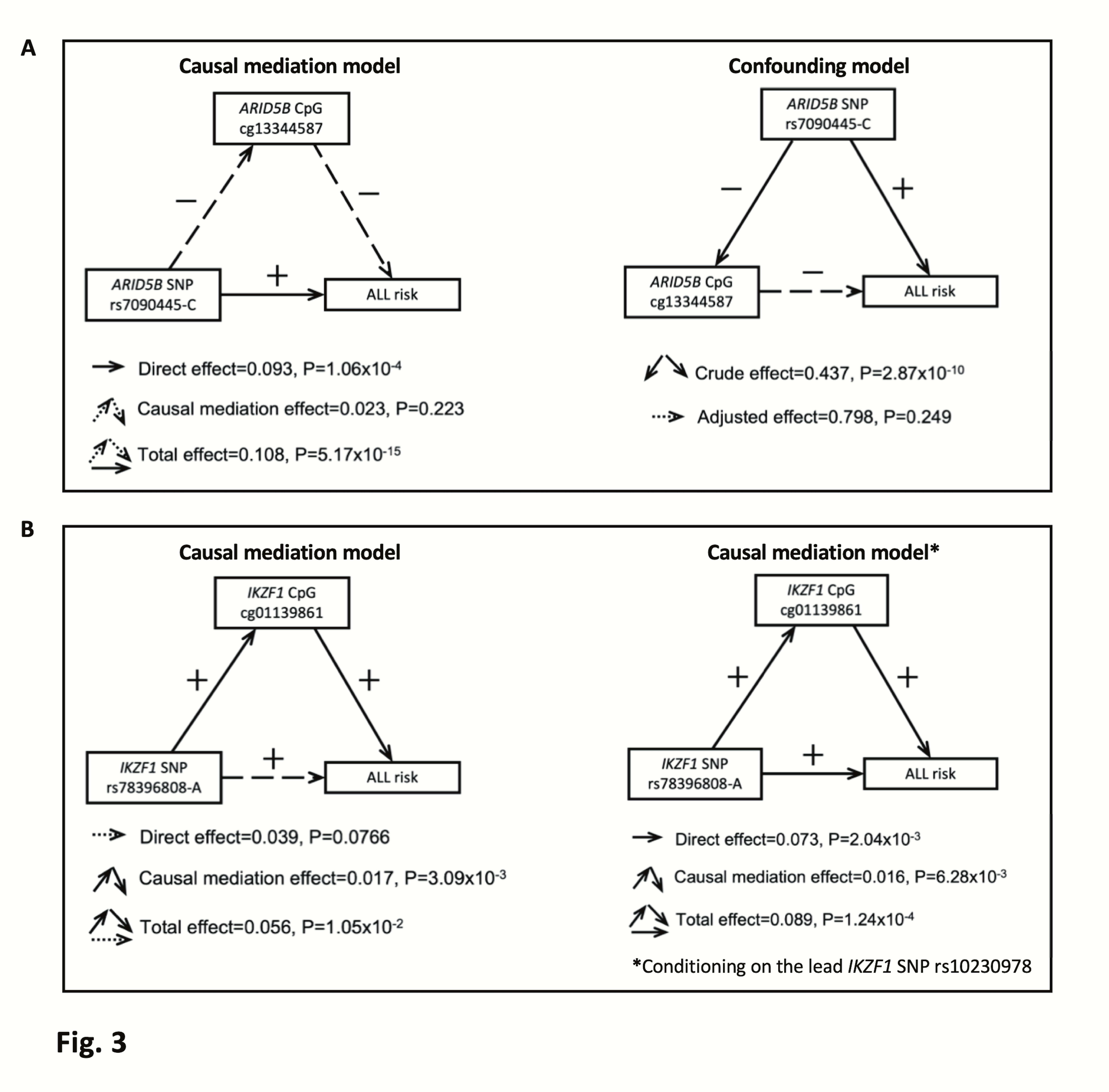
Causal mediation analysis¶
This is a workflow to perform model-based causal mediation analyses for the significant mQTL–DMP pairs, using the ‘mediation’ R package. First, we specified two statistical models, (i) the mediator model for the distribution of the DMP methylation level, after conditioning on the genotype of the mQTL and covariates including sex, ancestry, batch effect and cell type heterogeneity and (ii) the outcome model for the conditional distribution of ALL status, given the mQTL genotype, DMP methylation level and the same covariates. Models were fitted separately, and then, their fitted parameters were used as the main inputs to the mediate function, which computes the estimated ACME, the ADE and the total effect. Variances were estimated based on simulation. The quasi-Bayesian Monte Carlo simulation based on normal approximation was conducted 1000 times. Alternatively, an approach based on non-parametric bootstrap was also applied to estimate variance for validation. Results of the mediation analysis performed separately for three datasets were summarized across all the datasets using the fixed-effect meta-analysis model.
Study publication: link
Mediation script: link
Meta-analysis of mediation results link
Path diagrams showing the results of the causal mediation and confounding analyses: